Shimeld Lab Research
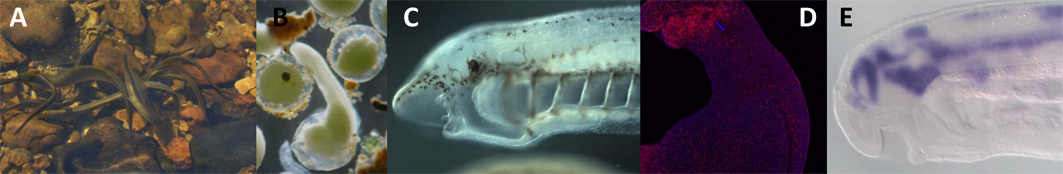
Animals come in a huge diversity of body forms. We seek to understand the evolutionary origins of this diversity, using tools drawn from ecology, developmental biology, evolutionary biology, molecular biology, genomics and bioinformatics. For an introduction to some of the species we work with, see my Favourite Animals section. Below describes some of the core research questions, with links to the relevant publications detailing our findings. Scroll down or link via the list below:
- Vertebrate origins: genes, organs, tissues and cell types
- Axes, symmetry and asymmetry
- Gene diversification in animal evolution
- Other projects: thermotolerance in marine larvae; the evolution of shells; microRNAs in animal evolution; developmental biology of colonial ascidians
Vertebrate origins: genes, organs, tissues and cell types
At both the genetic and morphological level vertebrates seem to be a lot more complicated than their invertebrate relatives. Vertebrates appear to have invented a number of new tissues, such as the neural crest and placodes, and to have massively elaborated others, such as the brain and spinal cord. They also have more regulatory genes than their invertebrate relatives, with most families of transcription factor and signalling molecule genes having expanded by gene duplication during early vertebrate evolution. We are investigating how both molecular and morphological complexity has evolved. Using molecular and embryological methods we are characterising the development of animals such as ascidians, amphioxus and lampreys, lineages of living creatures which span the origin of vertebrates. A special emphasis is placed on unravelling the changing roles of regulatory gene networks involved in the development of vertebrate specific morphology. Within this broad area several specific research projects are underway, including:
- The evolution of the vertebrate lens and crystallin genes
- The evolutionary origin of placodes and ganglia
- Transcriptomic profiling of sensory cells and the evolution of new cell types
- The origins of complex neural patterning
Publications in this area
Most living animals are members of the Bilateria and are, as the name suggests, at least superficially bilaterally symmetrical. Establishing bilateral symmetry follows from establishing definitive AP and DV axes, with midline cells often taking on an organising role. However the embryos of many species also deliberately break bilateral symmetry, forming asymmetric bodies with definitive if subtle left and right sides: the asymmetric placement of our own internal organs, and the spiral coiling of snail shells are examples of this.
We are using a variety of species (mollusc, annelid, amphioxus, lamprey) to dissect the evolution of the mechanisms that generate distinct left and right sides. We use confocal, EM and timelapse imaging to follow the initial breaking of symmetry during early cleavage, gene and signalling pathway manipulation to understand how this is transferred to asymmetric gene expression, and the tools of developmental biology to establish how such molecular asymmetry is then propagated.
Publications in this area
Gene diversification in animal evolution
The duplication of genes is of common occurrence in evolution. For example early vertebrate evolution is marked by widespread gene duplication resulting from duplication of the entire genome. We are using molecular phylogenetic approaches, coupled with genome sequencing of key taxa, to reconstruct the evolution of complex gene families. Such approaches allow us to ask when gene duplications have occurred, and just as importantly when genes have been lost and how orthologous genes have diverged in different lineages. We focus on ‘developmental’ genes, that is those involved in transcriptional control, intercellular signalling and tissue integrity, mapping changes onto phylogeny and allowing the construction of testable hypotheses of the underlying genetic basis for macroevolutionary change.
Publications in this area
Through a combination of collaboration and exploitation of the biological, genomic and transcriptomic resources we have developed we are also investigating a number of other aspects of invertebrate evolution and development. Please see the publications below or contact me if you want to know more about any of these areas.
Publications in these areas




